In this post I will present a technique for generating a one dimensional (quasi) fractal data set using a modified Matérn point process, perform a simple box-couting procedure, and then calculate the lacunarity and fractal dimension using linear regression. Lacunarity is actually a pretty large topic, and we will only cover one accepted interpretation here. This material was motivated by an interesting paper on the fractal modelling of fractures in tight gas reservoirs. Tight gas reservoirs refer to reservoirs with very low permeability. To provide a sense of perspective, oil reservoirs typically have a permebility of ten to a hundred millidarcies, whereas shale gas reservoirs are usually less than 0.1 microdarcies, which is about the same permeability as a granite countertop.
Fractals
The salient characteristic of fractals is their self similarity at multiple scales. My favorite analogy is that of a tree in winter. A stick with lots of twigs looks like a miniature branch, and a branch with lots of sticks and twigs looks like a miniature tree. There’s self similarity at (sort of) three different scales. If you’re looking up through the branches, the fractal dimension (kind of) describes how thickly the branches blot out the sky, and the lacunarity (sort of) describes the nature of the gaps between the branches. A larger lacunarity means bigger gaps, and a smaller lacunarity means smaller gaps. In fractal fracture modeling, the lacunarity is related to the density of the formation.
Lacunarity
Another way to think of lacunarity is to think of a map with lakes on it, as lacunarity derives from the Latin lacuna for gap or lake. (Think also of Laocoön, Poseidon’s priest in the Illiad.) Then a map with lots of lakes, like that of Minnesota, has a great degree of lacunarity, whereas a map of Arizona has substantially less so. That’s the four inch paint brush version. Lacunarity is a big concept with multiple interpretations. Here is a good site that has a more examples of lacunarity, and some more code for analyzing data with fractal analysis.
Fractal Dimension
Fractal dimension is a little bit easier to explain. If you think of a point in space as having zero dimension, and a line as having one dimension, then we can thing of an infinite cloud of points lying in a line as having a dimension somewhere between zero in one. Likewise, if a we think of a solid plane as having two dimensions, then we can imagine an infinitely fine mesh as having somewhat less than two whole dimensions, as it is not solid through and through, but more than one dimension as it has a width greater than a line. We can imagine other things, like infinitely spongy sponges in three dimesnions, and then for most of us our imaginative capacities totally fail us when we hit four dimensions, but three is good enough to get the general idea.
The Box Counting Algorithm
The idea of the box algorithm is to subdivide the domain into boxes, and count the number of boxes that contain observations of the data. For example, if your domain is the segment ![]() , your data is
, your data is ![]() , and you split your domain into four equal sized boxes, then you have a box count of 3, because none of your data falls into the
, and you split your domain into four equal sized boxes, then you have a box count of 3, because none of your data falls into the ![]() box. (The endpoints of the boxes are an “implementation detail”.) Once you complete one box counting, you double the number of boxes, thereby halving their sizes, and take another count. What you’re measuring is the number of boxes needed to cover or contain a fractal, as a function of box size or resolution. This relationship between the number of boxes and the box size is then plotted on a log-log plot. If the data has a multiresolution structure, then the box counting algorithm will produce a roughly linear log-log plot. We can then fit a line to this plot and take its slope and intercept as the fractal dimension and the lacunarity, respectively.
box. (The endpoints of the boxes are an “implementation detail”.) Once you complete one box counting, you double the number of boxes, thereby halving their sizes, and take another count. What you’re measuring is the number of boxes needed to cover or contain a fractal, as a function of box size or resolution. This relationship between the number of boxes and the box size is then plotted on a log-log plot. If the data has a multiresolution structure, then the box counting algorithm will produce a roughly linear log-log plot. We can then fit a line to this plot and take its slope and intercept as the fractal dimension and the lacunarity, respectively.
Word of Warning
These measures of fractal dimension and lacunarity are very, very rough. Two radically different looking fractals can have the exact same fractal dimension and lacunarity. Moreover, the calculation for lacunarity presented here is only one interpretation of lacunarity. Whereas a line can be described by a point and a slope, and a unimodal data set can usually be described by its mean and standard deviation, fractals and other similar data sets have much greater complexity and require a more lengthy description than a few scalar summary statistics.
Data
The data we will generate is not fractal in the popular mathematical sense, which is to say, it is not generated by an iterated function system, and it does not look like a Koch snowflake, a Barnsley fern, a Hilbert curve, a Mandelbrot set, or any other of your favorite fractals. It does however have a sort of self similarity at multiple scales. We will produce a Matérn process with four layers of (invisible) parents before collecting the child points. This will produce scattered dense multiresolution clusters that are reminiscent of fractals.
First we will import some modules,
import numpy as np, scipy, scipy.stats from pandas import Series, DataFrame import scipy.optimize
Next we’ll generate some data. We’ll begin by picking a N of parent points uniformly from the domain ![]() , where
, where ![]() . We write a function to generate a number of child points according to a Matérn point process. We then apply that function over several generations, producing many children.
. We write a function to generate a number of child points according to a Matérn point process. We then apply that function over several generations, producing many children.
L = 64
U = list()
rate = L / 6.0
N = scipy.stats.poisson( rate ).rvs()
U = list( scipy.stats.uniform(0,L).rvs(N) )
def matern_children( U, rate, nbhd ):
T = list()
for i in range( len( U ) ):
M = scipy.stats.poisson( rate ).rvs()
for j in range( M ):
T.append( scipy.stats.uniform( U[i]-nbhd, 2*nbhd ).rvs() )
return T
nbhd = 2.0
U = matern_children( U, rate, nbhd )
U = matern_children( U, rate, nbhd/4.0 )
U = matern_children( U, rate, nbhd/8.0 )
U = matern_children( U, rate, nbhd/16.0 )
For each generation, we shrink the neighborhood for the children while keeping the Poisson rate the same. This produces smaller and smaller bunches of children in each generation. We can then see our distribution by using a histogram. We see that his process produces a data set that is either empty, or densely clustered.
hist( U, 100, alpha=0.5 ) ;
title('Distribution of Four Tier Matern Point Process')
ylabel('Frequency') ; xlabel('Location') ;
savefig('matern_fractal_distribution.png', fmt='png', dpi=100 )

Implementing the Box Counting Algorithm
This implementation is provided as an example only. It probably would not scale well to a two dimensional problem, let alone a highly dimensional data set. I have used the R package fdim for research, but I am not sure if that is the best (free) tool available.
def count_boxes( data, box_size, M ):
data = Series( data )
N = np.int( np.floor( M / box_size ) )
counts = list()
for i in range( N ):
condition = ( data >= i*box_size )&( data < (i+1)*box_size )
subset = data[ condition ]
counts.append( subset.count() )
counts = [ i for i in counts if i != 0 ]
return len( counts )
r = np.array([ L/(2.0**i) for i in range(12,0,-1) ])
N = [ count_boxes( U, ri, L ) for ri in r ]
Here, r is an array of box lengths, and N is the number of boxes needed to cover the fractal for a given box length. We will plot N against 1./r on a log-log plot so that we’ll have a positive linear relationship, but before we plot anything, we will model the data using the curve_fit() function from scipy.optimize.
Modeling
If we assume that there is a power law relationship between the box size ![]() , and the number of boxes
, and the number of boxes ![]() required to cover the fractal, then we can write,
required to cover the fractal, then we can write,
(1) ![]()
Then if we take the log of both sides we obtain,
(2) ![]()
And now we have a nice linear model for our box counting data where A represents the lacunarity, and Df represents the fractal dimension. We can encode this model in the following function f(x,A,Df) for use in the curve_fit() optimization routine which will attempt to find optimal values for A and Df.
def f( x, A, Df ):
'''
User defined function for scipy.optimize.curve_fit(),
which will find optimal values for A and Df.
'''
return Df * x + A
popt, pcov = scipy.optimize.curve_fit( f, np.log( 1./r ), np.log( N ) )
A, Df = popt
The function curve_fit() accepts a model, and the empirical input and output values, and then returns the optimized parameters in the variable popt, and an estimate of the variance and covariance of those parameters in pcov. We’ll extract the lacunarity A and the fractal dimension Df.
fig, ax = subplots()
plot( 1./r, N, 'b.-' )
ax.plot( 1./r, np.exp(A)*1./r**Df, 'g', alpha=1.0 )
ax.set_xscale('log')
ax.set_yscale('log')
ax.set_aspect(1)
xlabel('Box Size, 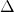 ')
ylabel('Number of Boxes,
')
ylabel('Number of Boxes, 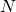 ')
grid(which='minor', ls='-', color='0.75')
grid(which='major', ls='-', color='0.25')
title('Box-Counting')
savefig('fractal_box_counting.png',fmt='png',dpi=100)
')
grid(which='minor', ls='-', color='0.75')
grid(which='major', ls='-', color='0.25')
title('Box-Counting')
savefig('fractal_box_counting.png',fmt='png',dpi=100)

This graph should make intuitive sense. For large box sizes ![]() , or very small
, or very small ![]() , we shouldn’t need that many boxes to cover the fractal. For smaller box sizes
, we shouldn’t need that many boxes to cover the fractal. For smaller box sizes ![]() , or very large
, or very large ![]() , we should expect to need very many boxes to cover a fractal. This example is particularly nice because it is an artificial data set and we can zoom through many scales. In practice, one is often limited by the resolution of a sensor and the log-log plot may come up sharply at the beginning and hit a plateau at the end at the limit of the resolution of the sensor. In this case, one might only have a handful of points in the middle for a linear regression.
, we should expect to need very many boxes to cover a fractal. This example is particularly nice because it is an artificial data set and we can zoom through many scales. In practice, one is often limited by the resolution of a sensor and the log-log plot may come up sharply at the beginning and hit a plateau at the end at the limit of the resolution of the sensor. In this case, one might only have a handful of points in the middle for a linear regression.
Another Modeling Approach
I wanted to demonstrate the curve_fit() function because it is a really neat tool, but we can also use statsmodels to perform linear regression. The advantage to this approach is that we can see the effect of trimming the very large and very small box sizes to obtain a better linear regression. Notes on how to interpret that statistics in the table below can be found in this previous post.
import statsmodels.formula.api as sm Y = np.log( N ) X = np.log( 1./D ) T = np.vstack((Y,X,np.ones_like(X))).T df = DataFrame( T, columns=['N(r)','Df','A'] ) Y = df['N(r)'] X = df[['Df','A']] result = sm.OLS( Y, X ).fit() result.summary()
I’ve highlighted the values in the table for the fractal dimension, Df, and the lacunarity, A.
OLS Regression Results
| Dep. Variable: | N(r) | R-squared: | 0.998 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.997 |
| Method: | Least Squares | F-statistic: | 4178. |
| Date: | Tue, 04 Mar 2014 | Prob (F-statistic): | 1.91e-14 |
| Time: | 20:33:55 | Log-Likelihood: | 9.9843 |
| No. Observations: | 12 | AIC: | -15.97 |
| Df Residuals: | 10 | BIC: | -15.00 |
| Df Model: | 1 |
| coef | std err | t | P>|t| | [95.0% Conf. Int.] | |
|---|---|---|---|---|---|
| Df | 0.8995 | 0.014 | 64.640 | 0.000 | 0.869 0.931 |
| A | 3.7758 | 0.034 | 112.222 | 0.000 | 3.701 3.851 |
| Omnibus: | 2.101 | Durbin-Watson: | 0.590 |
|---|---|---|---|
| Prob(Omnibus): | 0.350 | Jarque-Bera (JB): | 0.946 |
| Skew: | 0.192 | Prob(JB): | 0.623 |
| Kurtosis: | 1.679 | Cond. No. | 2.45 |
Very helpful.
Thanks
in X = np.log( 1./D ), what is D ?